General areas of research interest:
Molecular Microbiology, Bacterial Genetics, Regulation of Gene Expression, Nutrient uptake, Synthetic Biology (iGEM)
Just because bacteria are thought to be the simplest living organisms, they are by no means “simple”! When a single bacterium divides by binary fission, the result is two daughter cells that are genetically identical. But for a long time, scientists assumed that the two daughter cells would also have identical shapes and cellular contents (proteins, mRNAs, etc.) However, we now know that for many species, the two daughter cells have very distinct shapes, distinct contents, and distinct abilities from each other. So, how can this differentiation happen? It depends on precisely controlled gene expression and protein localization
so that the right proteins are in the right places at the right times. Which proteins are involved? What do they do? Where are they located in each of the daughter cells? When are they expressed? How is their activity controlled? These are the questions I’m interested in and they’re also some of the most fundamental questions in developmental biology.
Scope of Research Projects:
My research involves several independent, but related projects and I encourage students to meet with me if they want to learn more. I’ll give you material to read and think about and then craft a project with you that meets your goals. I have small projects that could be completed in a semester by an individual student and long-term projects for students who want to start as a freshman or sophomore and carry out a project throughout their undergraduate education. I encourage students to come to me with ideas as well.
Model Organism:
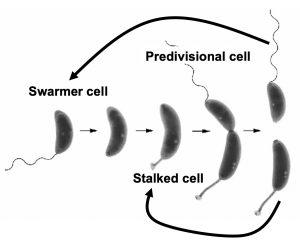
The organism I use to investigate these questions is the aquatic bacterium, Caulobacter crescentus, which flourishes in low-nutrient environments such as rivers, lakes, oceans, and even bottled water. In order to survive in its competitive environment, Caulobacter undergoes a unique life cycle. Each cell division produces two morphologically distinct daughter cells: the swarmer cell has a flagellum that propels it through the water, allowing it to locate sparse nutrients, and the stalked cell has a protrusion of its cell envelope (a stalk) containing an extremely sticky substance at the tip that cements it to surfaces. After cell division, the stalked cell immediately begins a new round of chromosome replication and division, but the swarmer cell must take the time to differentiate into a stalked cell before it can initiate DNA replication (Fig. 1). This complex cell division cycle is controlled by a fascinating network of signal transduction proteins, some of which are well-understood, and some of which have yet to be discovered or well-characterized.
Caulobacter crescentus is a great organism to use in the lab. It’s relatively easy to obtain a culture that consists only of swarmer cells and then observe the entire population of cells move in synchrony through the different stages of the cell cycle. When the cells are synchronized, we can study which proteins are expressed at which times of the cell cycle and where those proteins are located within each type of cell. Caulobacter is non-pathogenic, aerobic, and grows well on defined medium in a laboratory. With Caulobacter, we can perform many classical genetic techniques such as genetic screens for suppressor mutations, phage transductions, conjugation, etc. Its genome has been sequenced, which allows us to use bioinformatics to identify genes of interest and compare them with genes in other sequenced organisms. Also, Caulobacter is “genetically tractable” so we can use many molecular genetic techniques such as gene knock-outs, induction or repression of gene expression, and gene fusions (we often tag our proteins with Green Fluorescent Protein (GFP) to see where they are located within the cell). We also use imaging techniques such as fluorescent and DIC microscopy and biochemical techniques such as Western Blots, gel shift assays, enzyme activity assays, co-immunoprecipitations and more.