** Joiner lab members are bolded.
Joiner, C.M., Glogowski, T.J., NewRingeisen, E.M., Huynh, H.V., Roberts, M.G., Rognerud, M.M., Huebsch, H.E. Photoactivatable O-GlcNAc transferase library enables covalent chemical capture of solvent-exposed TPR domain interactions. ChemBioChem 2024, 26, e202400709
Listenberger, L.L., Joiner, C.M., Terrell, C.R. Using open-source bioinformatics and visualization tools to explore the structure and function of SARS-CoV-2 spike protein. CourseSource, 2022.
Postdoctoral Work:
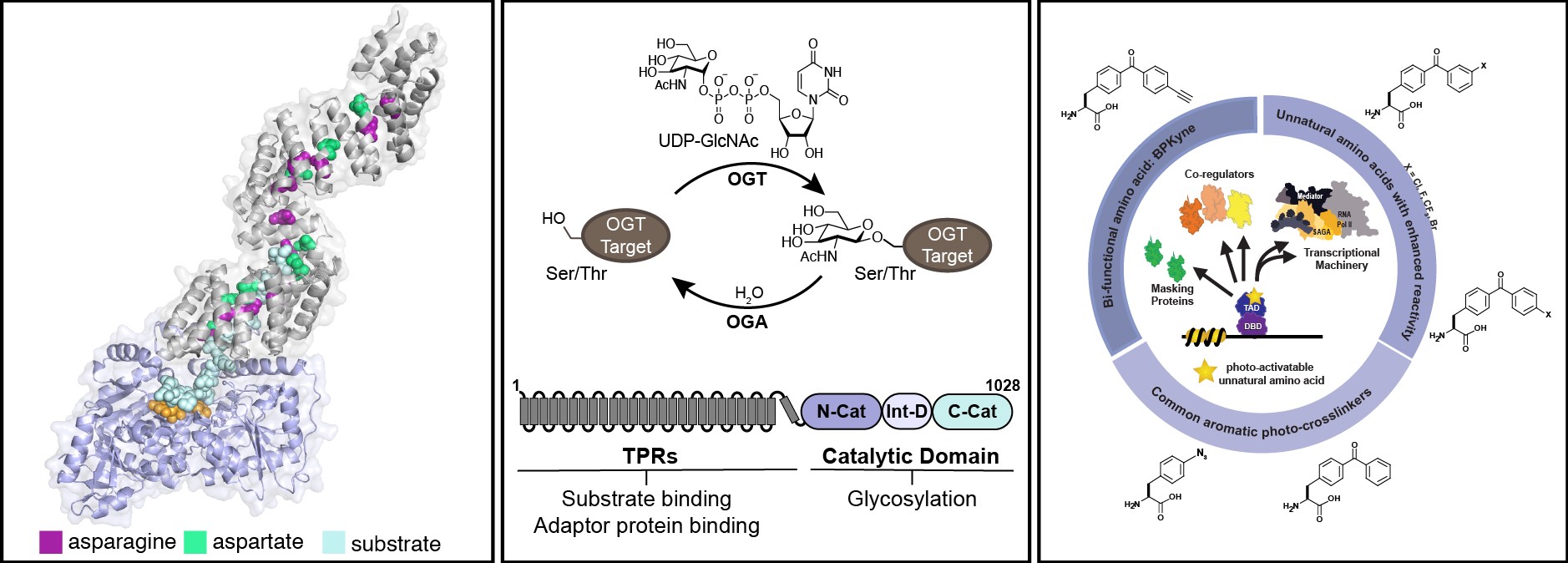
Potter, S.C., Gibbs, B.E., Hammel, F.A., Joiner, C.M., Paulo J.A., Janetzko, J., Levine, Z.G., Fei, G.Q., Haggarty, S.J., Walker, S. Dissecting OGT’s TPR domain to identify determinants of cellular function. PNAS 2024, 121, 22, e2401729121.
Joiner, C.M., Hammel, F.A., Janetzko, J., Walker, S. Protein substrates engage the lumen of O-GlcNAc transferase’s tetratricopeptide repeat domain in different ways. Biochemistry 2021, 60, 11, 847-853.
Levine, Z.M., Potter, S.C., Joiner, C.M., Fei, G.Q., Nabet, B., Sonnett, M., Zachara, N.E., Gray, N.S., Paulo, J.A., Walker, S. Mammalian cell proliferation requires noncatalytic functions of O-GlcNAc transferase. Proc. Natl. Aca. Sci. USA. 2021, 118, 4, e2016778118
Joiner, C.M.*, Levine, Z.M.*, Aonbangkhen, C., Woo, C.M., Walker, S. Aspartate residues far from active site drive O-GlcNAc transferase substrate selection. Journal of American Chemical Society 2019, 141, 33, 12974-12978. *denotes co-first authors
Joiner, C.M., Li, H., Jiang, J., Walker, S. Structural characterization of the O-GlcNAc cycling enzymes: insights into substrate recognition and catalytic mechanisms. Current Opinions in Structural Biology 2019, 56, 97-106.
Graduate Work:
Joiner, C.M.*, Breen, M.E.*, Mapp, A.K. Electron-deficient p-benzoyl-L-phenylalanine derivatives increase covalent chemical capture yields for protein-protein interactions. Protein Science 2019, 28(6), 1163-1170. *denotes co-first authors
Joiner, C.M.*, Breen, M.E.*, Clayton, J., Mapp, A.K. A bifunctional amino acid enables both covalent chemical capture and isolation of in vivo protein-protein interactions. ChemBioChem 2017, 18(2), 181-184. *denotes co-first authors
Lancia, J., Nwokoye, A., Dugan, A., Joiner, C., Pricer, R., Mapp, A. Sequence context and crosslinking mechanism affect the efficiency of in vivo capture of protein-protein interactions. Biopolymers 2014, 101(4), 391-397.